Flowcharts of the Models Logic
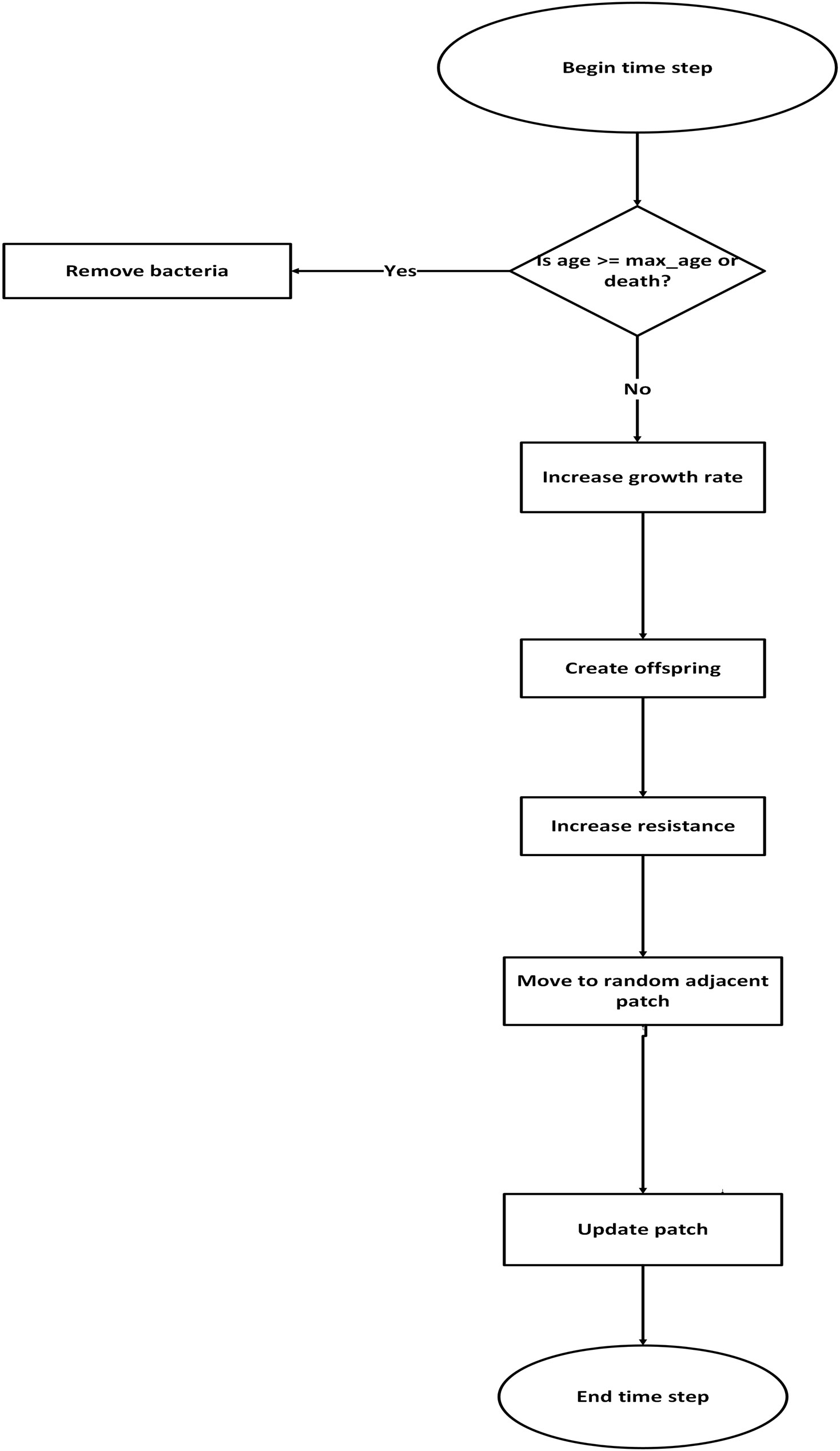
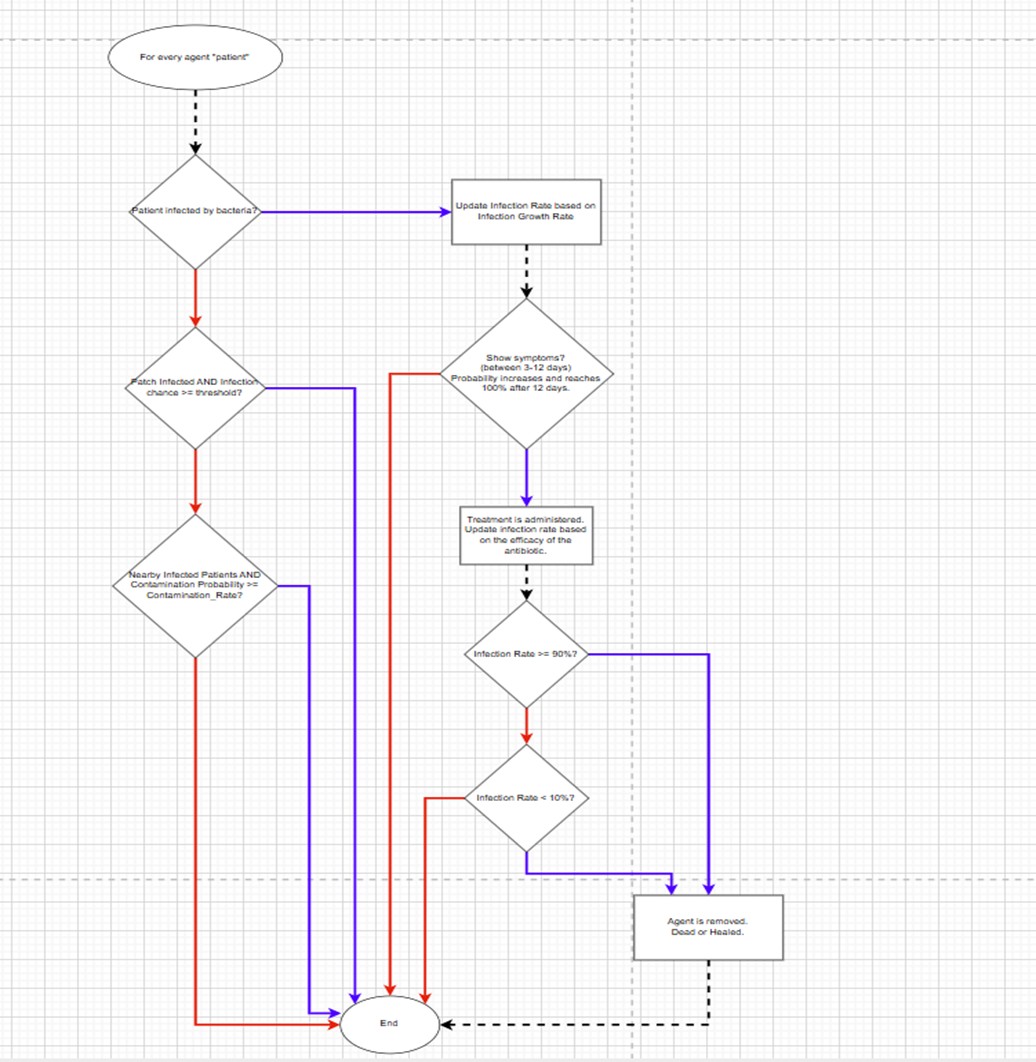
These flowcharts summarize the core logic of our NetLogo model: the left diagram outlines bacteria lifecycle events (growth, mutation, death, and environmental cleaning), and the right diagram shows transmission pathways across human agents (infection, treatment, recovery, and discharge). Together, they provide a high‑level view of how parameters influence outcomes during simulation.
Code
NetLogo Model
globals [
time-step
total-mutations
current-infected
total-infections
total-recovered
percent-total-infected
initial-population-per-species
successful-antibiotics
successful-cleans
patient-deaths
total-bacteria
total-discharged
]
breed [patients patient]
breed [bacteria bacterium]
patches-own [
patch-density-color
]
patients-own [
patient-id
age
immunocompromised?
infected?
pathogen-resistant?
antibiotic-timer
alive?
patient-status
days-infected
antibiotic-failures
healthy-ticks
ever-healed?
symptoms?
symptom-delay ;; ticks until symptoms appear (300–1200)
symptom-onset-tick ;; tick when symptoms started
antibiotics-after-symptoms-count ;; number of antibiotic administrations after symptoms
]
bacteria-own [
species
growth-rate
death-rate
carbapenem-resistant?
bacteria-age
]
to setup
clear-all
set-default-shape patients "person"
set-default-shape bacteria "dot"
set time-step 0
set initial-population-per-species 10
;; Initialize Counters
set successful-antibiotics 0
set successful-cleans 0
set patient-deaths 0
setup-environment
setup-patients
setup-bacteria
reset-ticks
end
to setup-environment
ask patches [
set pcolor white
set patch-density-color white
]
end
to setup-patients
create-patients total-patients [
set size 1.3
setxy random-xcor random-ycor
init-patient-vars
]
end
to init-patient-vars
set patient-id who
set age random 90
set immunocompromised? (random-float 1 < 0.2)
set infected? false
set pathogen-resistant? false
set antibiotic-timer 0
set alive? true
set days-infected 0
set patient-status "healthy"
set color green
set antibiotic-failures 0
set healthy-ticks 0
set ever-healed? false
set symptoms? false
set symptom-delay 0
set symptom-onset-tick -1
set antibiotics-after-symptoms-count 0
end
to setup-bacteria
let species-list ["Acinetobacter baumannii" "Enterobacteriaceae" "Pseudomonas aeruginosa"]
foreach species-list [
this-species ->
create-bacteria initial-population-per-species [
set size 0.5
setxy random-xcor random-ycor
set species this-species
set bacteria-age random 10
set carbapenem-resistant? (random-float 1 < 0.04)
if species = "Acinetobacter baumannii" [
set color ifelse-value carbapenem-resistant? [violet] [orange]
]
if species = "Enterobacteriaceae" [
set color ifelse-value carbapenem-resistant? [red] [pink]
]
if species = "Pseudomonas aeruginosa" [
set color ifelse-value carbapenem-resistant? [blue] [cyan]
]
; Adjusted rates to ensure survival between cleanings
set growth-rate 0.4
set death-rate 0.4
]
]
end
to go
;; Ensure batch run rules enforced
;; Testing
;; Stop if total bacteria over 5000 agents ;;
if total-bacteria > 5000 [ stop ]
;; Stop if over 80% patches contaminated
if (count patches with [count bacteria-here > 0] / count patches) > 0.8 [
stop
]
;; Stop if all patients are dead
if not any? patients with [alive?] [ stop ]
;; End testing ;;
;; Stop if no bacteria present
if not any? bacteria [ stop ]
;; Continue simulation if none of the stop conditions are met
set time-step time-step + 1
;; Patient Spawning Logic
if random-float 1 < 0.10 [
let empty-patches patches with [not any? patients-here]
if any? empty-patches [
create-patients 1 [
move-to one-of empty-patches
set size 1.3
init-patient-vars
]
]
]
ask patients [ patient-behavior ]
ask bacteria [ bacteria-behavior ]
ask patches [ environment-behavior ]
update-globals
tick
end
to patient-behavior
if not alive? [ stop ]
ifelse infected? [
set patient-status "infected"
set color red
set days-infected days-infected + 1
set antibiotic-timer antibiotic-timer + 1
set healthy-ticks 0 ; reset since patient is sick
;; Symptoms appear between 300 and 1200 ticks post-infection
;; Immunocompromised patients show symptoms sooner (bias to shorter window)
if (not symptoms?) [
if (symptom-delay = 0) [
ifelse immunocompromised? [
set symptom-delay 250 + random 801 ;; ~3–10.5 days
] [
set symptom-delay 300 + random 901 ;; ~3–12 days
]
]
if (days-infected >= symptom-delay) [
set symptoms? true
set symptom-onset-tick ticks
]
]
;; Allow antibiotics only after symptoms
if (antibiotic-application and symptoms?) [
if antibiotic-timer >= antibiotic-administration-period [
set antibiotic-timer 0
administer-antibiotics
set antibiotics-after-symptoms-count antibiotics-after-symptoms-count + 1
]
]
;; Death condition: 700 ticks after symptoms if at least one antibiotic was administered and not cured
if (symptoms? and (antibiotics-after-symptoms-count >= 1) and ((ticks - symptom-onset-tick) >= 700)) [
set alive? false
set patient-deaths patient-deaths + 1
die
]
] [
;; Healthy patient logic
check-for-environment-infection
set patient-status "healthy"
set color green
;; Track consecutive healthy ticks
set healthy-ticks healthy-ticks + 1
;; Leave if healthy for over 100 ticks
if healthy-ticks >= 100 [
set total-discharged total-discharged + 1
die
]
]
end
to check-for-environment-infection
;; Patients who were healed by antibiotics cannot be reinfected
if ever-healed? [ stop ]
if count bacteria-here > 0 [
let base-prob 0.5
if immunocompromised? [ set base-prob base-prob + 0.2 ]
if random-float 1 < base-prob [
set infected? true
set color red
set antibiotic-timer 0
set days-infected 0
set symptoms? false
;; schedule symptoms window with immunocompromised bias
ifelse immunocompromised? [
set symptom-delay 250 + random 801
] [
set symptom-delay 300 + random 901
]
set symptom-onset-tick -1
set antibiotics-after-symptoms-count 0
let source-bacterium one-of bacteria-here
set pathogen-resistant? [carbapenem-resistant?] of source-bacterium
set total-infections total-infections + 1
]
]
end
to administer-antibiotics
let success-chance 0
;; Base success probability depends on bacterial resistance
ifelse pathogen-resistant? [
set success-chance 0.68 ; resistant baseline
] [
set success-chance 0.87 ; non-resistant baseline
]
;; Immunocompromised patients respond worse (-15%)
if immunocompromised? [
set success-chance success-chance - 0.15
]
;; Example: dosage=10 → +0.33, dosage=50 → +0.71, dosage=100 → +0.83
set success-chance success-chance + (antibiotic-strength-level / (antibiotic-strength-level + 20))
;; Clamp to [0, 1]
if success-chance > 1 [ set success-chance 1 ]
if success-chance < 0 [ set success-chance 0 ]
;; Roll for outcome
ifelse random-float 1 < success-chance [
;; SUCCESS
set infected? false
set pathogen-resistant? false
set color green
set patient-status "healed"
set successful-antibiotics successful-antibiotics + 1
set total-recovered total-recovered + 1
set antibiotic-failures 0
set ever-healed? true ; NEW: mark patient as immune to reinfection
] [
;; FAILURE
set antibiotic-failures antibiotic-failures + 1
if antibiotic-failures >= 3 [
set alive? false
set patient-status "dead"
set color black
set patient-deaths patient-deaths + 1
die
]
]
end
to bacteria-behavior
set bacteria-age bacteria-age + 1
rt random 360
fd 1
;; Mutation chance each tick
if random-float 1 < 0.01 [
if not carbapenem-resistant? [
set carbapenem-resistant? true
set total-mutations total-mutations + 1
;; Update color to resistant variant
if species = "Acinetobacter baumannii" [ set color violet ]
if species = "Enterobacteriaceae" [ set color red ]
if species = "Pseudomonas aeruginosa" [ set color blue ]
]
]
if time-step mod 10 = 0 [
; 30% chance to duplicate
if random-float 1 < growth-rate [
hatch 2 [
set species [species] of myself
set carbapenem-resistant? [carbapenem-resistant?] of myself
set color [color] of myself
set bacteria-age 0
]
die
stop
]
; 50% chance to die off
if random-float 1 < death-rate [
die
stop
]
]
end
to environment-behavior
; 1. CLEANING LOGIC (Only runs every 30 time steps)
if (time-step mod cleaning-frequency = 0) [
let num-bacteria count bacteria-here
if num-bacteria > 0 [
; Use slider value directly for kill percentage
let kill-percentage cleaning-effectiveness
; Check if this counts as a "successful clean" (> 70%)
if kill-percentage > 0.70 [
set successful-cleans successful-cleans + 1
]
; Calculate kill count
let kill-count floor (num-bacteria * kill-percentage)
ask n-of kill-count bacteria-here [ die ]
]
]
; 2. VISUALIZATION LOGIC (Runs every tick to show growth)
ifelse count bacteria-here = 0 [
set pcolor white
] [
set pcolor scale-color yellow count bacteria-here 0 30
]
end
to update-globals
; Calculate stats without overwriting slider variables
let current-population count patients
set current-infected count patients with [infected?]
if current-population > 0 [
set percent-total-infected (current-infected / current-population)
]
set total-bacteria count bacteria
end